Abstract
While molecular pre-training has shown great potential in enhancing drug discovery, the lack of a solid physical interpretation in current methods raises concerns about whether the learned representation truly captures the underlying explanatory factors in observed data, ultimately resulting in limited generalization and robustness. Although denoising methods offer a physical interpretation, their accuracy is often compromised by ad-hoc noise design, leading to inaccurate learned force fields. To address this limitation, this paper proposes a new method for molecular pre-training, called sliced denoising (SliDe), which is based on the classical mechanical intramolecular potential theory. SliDe utilizes a novel noise strategy that perturbs bond lengths, angles, and torsion angles to achieve better sampling over conformations. Additionally, it introduces a random slicing approach that circumvents the computationally expensive calculation of the Jacobian matrix, which is otherwise essential for estimating the force field. By aligning with physical principles, SliDe shows a 42\% improvement in the accuracy of estimated force fields compared to current state-of-the-art denoising methods, and thus outperforms traditional baselines on various molecular property prediction tasks.
Method
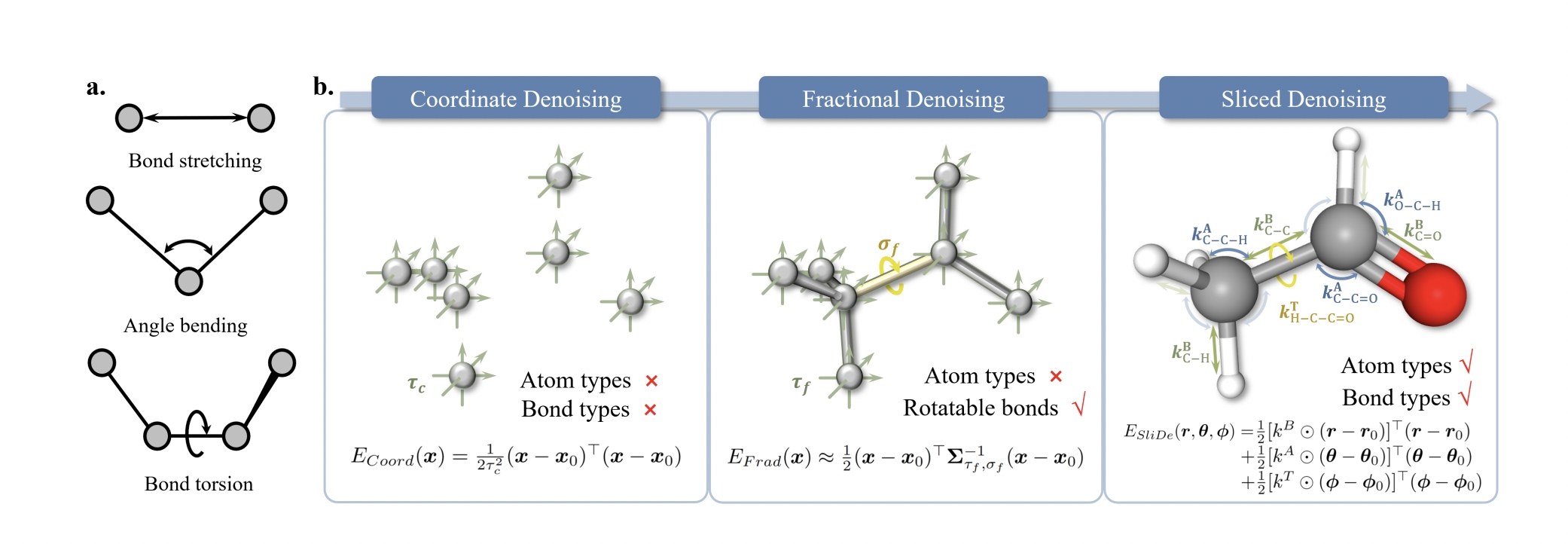
Comparison of the three denoising methods in terms of energy functions. Coordinate denoising learns an isotropic energy in Cartesian coordinates that does not discriminate different atom types and bond types. Based on coordinate denoising, fractional denoising treats the rotatable bonds in special. In contrast, sliced denoising performs fine-grained treatment for different atom types and bond types, enabling the most physically consistent description of the molecule.
Experiments
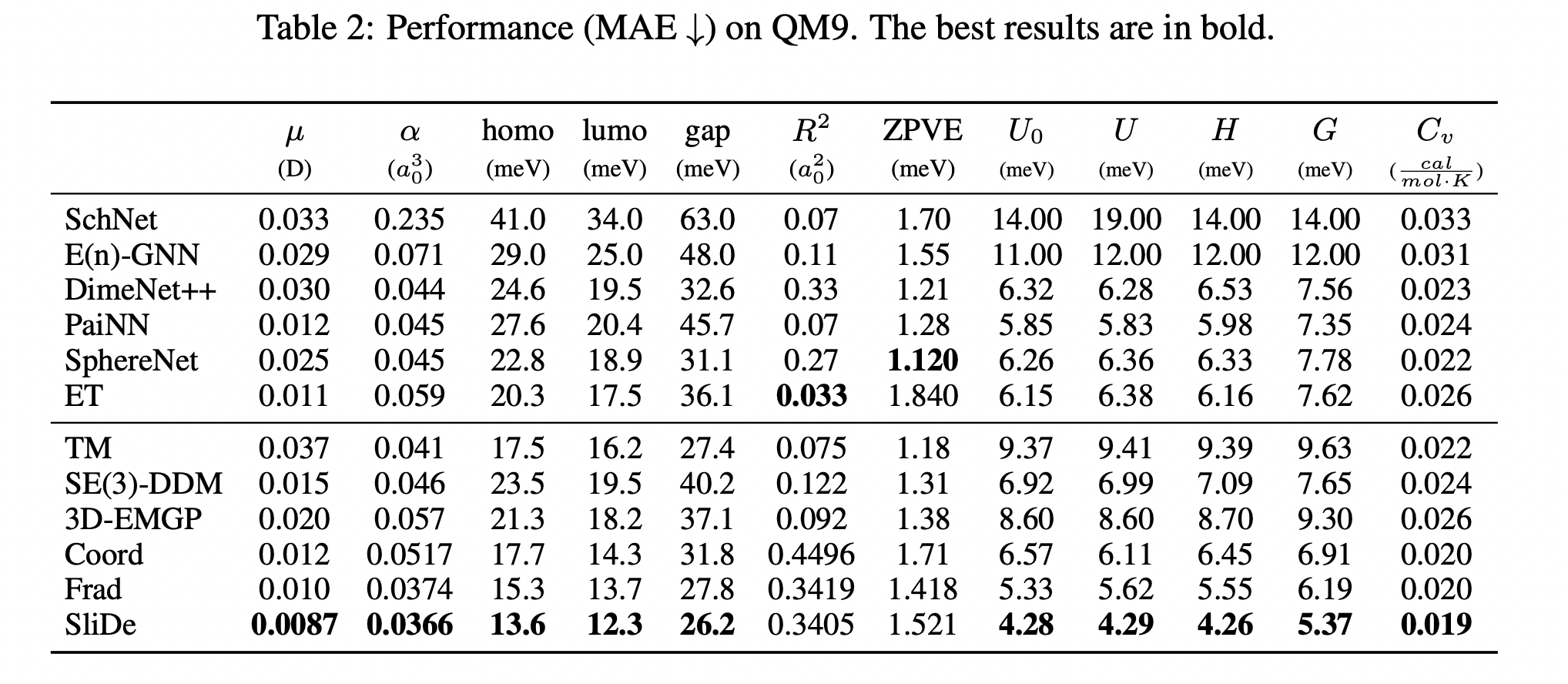
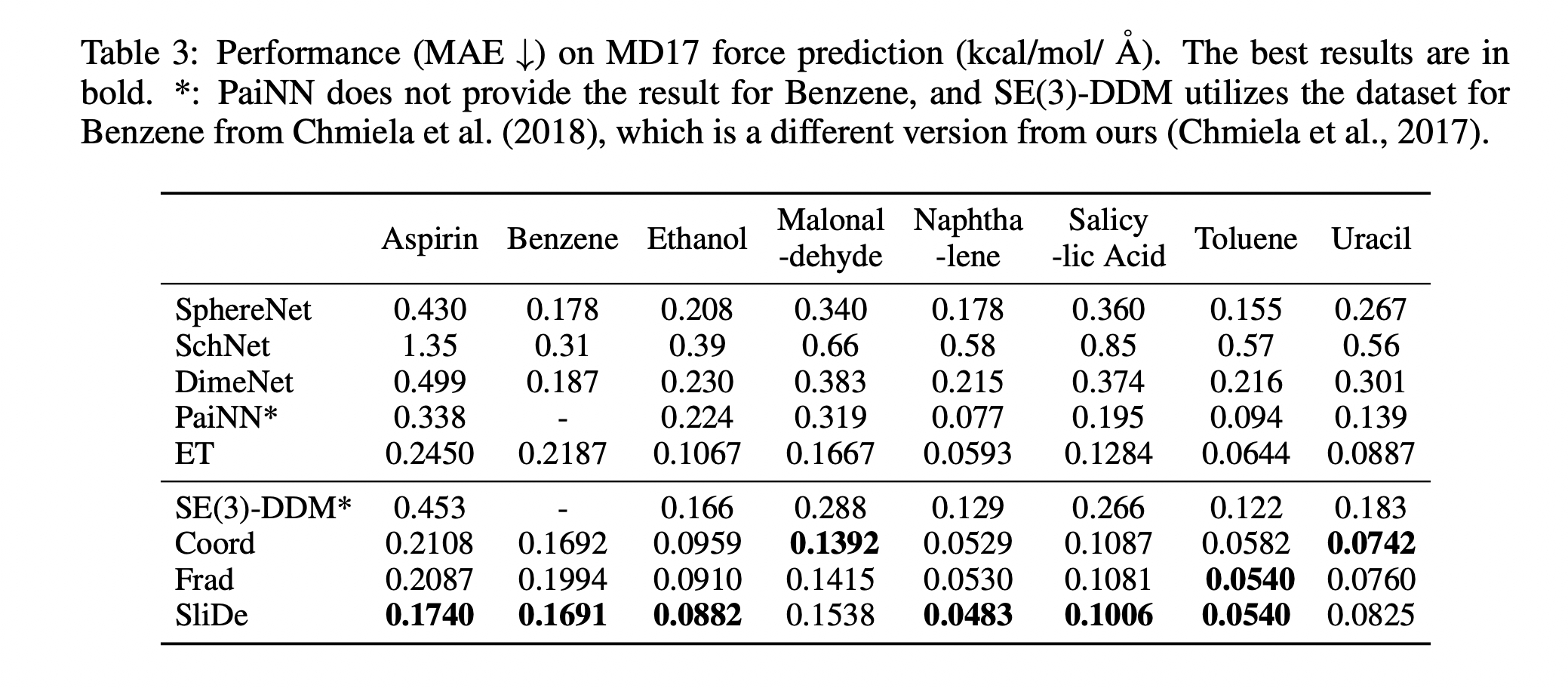
Performances (MAE, lower is better) on QM9 and MD17 force prediction are shown above. The best results are in bold.