Abstract
Pocket representations play a vital role in various biomedical applications, such as druggability estimation, ligand affinity prediction, and de novo drug design. While existing geometric features and pretrained representations have demonstrated promising results, they usually treat pockets independent of ligands, neglecting the fundamental interactions between them. However, the limited pocket-ligand complex structures available in the PDB database (less than 100 thousand non-redundant pairs) hampers large-scale pretraining endeavors for interaction modeling. To address this constraint, we propose a novel pocket pretraining approach that leverages knowledge from high-resolution atomic protein structures, assisted by highly effective pretrained small molecule representations. By segmenting protein structures into drug-like fragments and their corresponding pockets, we obtain a reasonable simulation of ligand-receptor interactions, resulting in the generation of over 5 million complexes. Subsequently, the pocket encoder is trained in a contrastive manner to align with the representation of pseudo-ligand furnished by some pretrained small molecule encoders. Our method, named ProFSA, achieves state-of-the-art performance across various tasks, including pocket druggability prediction, pocket matching, and ligand binding affinity prediction. Notably, ProFSA surpasses other pretraining methods by a substantial margin. Moreover, our work opens up a new avenue for mitigating the scarcity of protein-ligand complex data through the utilization of high-quality and diverse protein structure databases.
Method
Data Processing
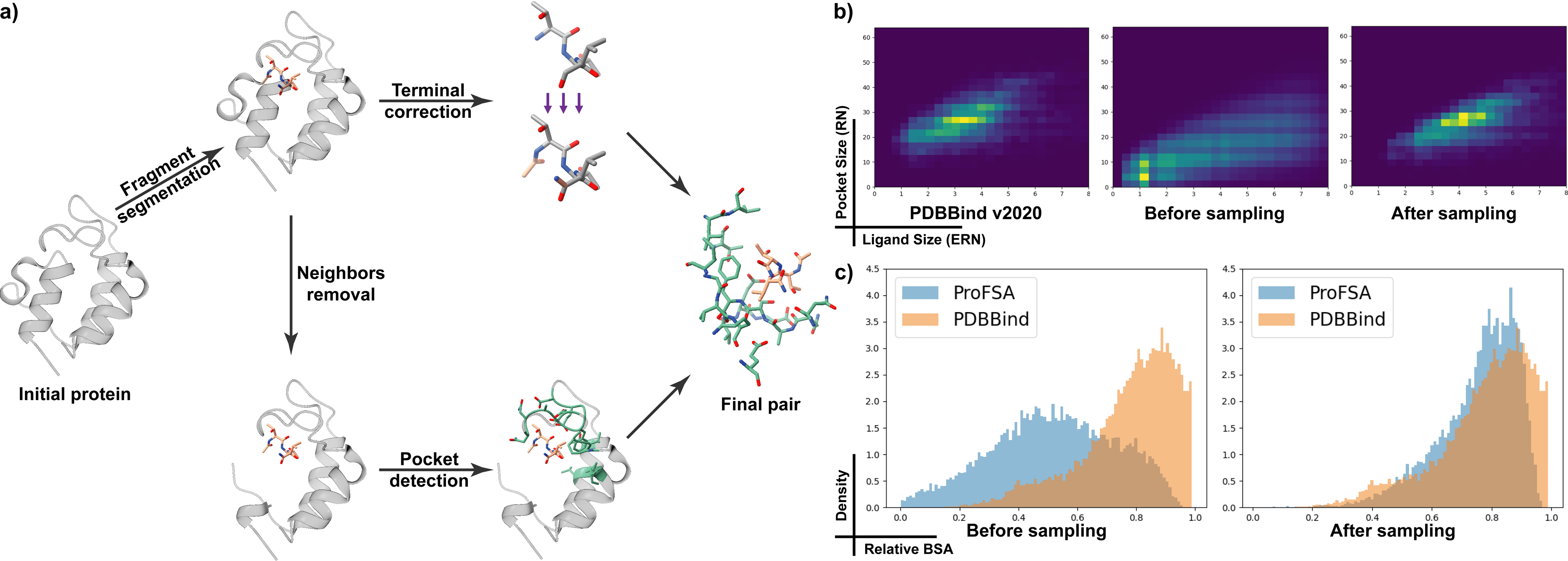
We iteratively isolate fragments from a protein structure and apply terminal corrections to fix biases introduced by breaking peptide bonds during fragment segmentation, resulting in the pseudo-ligands. Then, we exclude the five nearest residues on each side of the acquired fragment, focusing on long-range interactions. We then designate the surrounding residues containing at least one heavy atom within a {6\AA} proximity to the fragment as the pocket. This process yields pairs of finalized pockets and pseudo-ligands. The derived complexes are subjected to stratified sampling based on the PDBBind(v2020) distribution.
Training
Pockets are encoded by our pocket encoder, which is trained to align with fragment representations given by fixed pretrained molecule encoders. A simplified hydropathy-related (indicated by blue or orange color) example illustrates that fragment properties recognized by pretrained molecule encoders could guide pocket representation learning.

Experiment Results
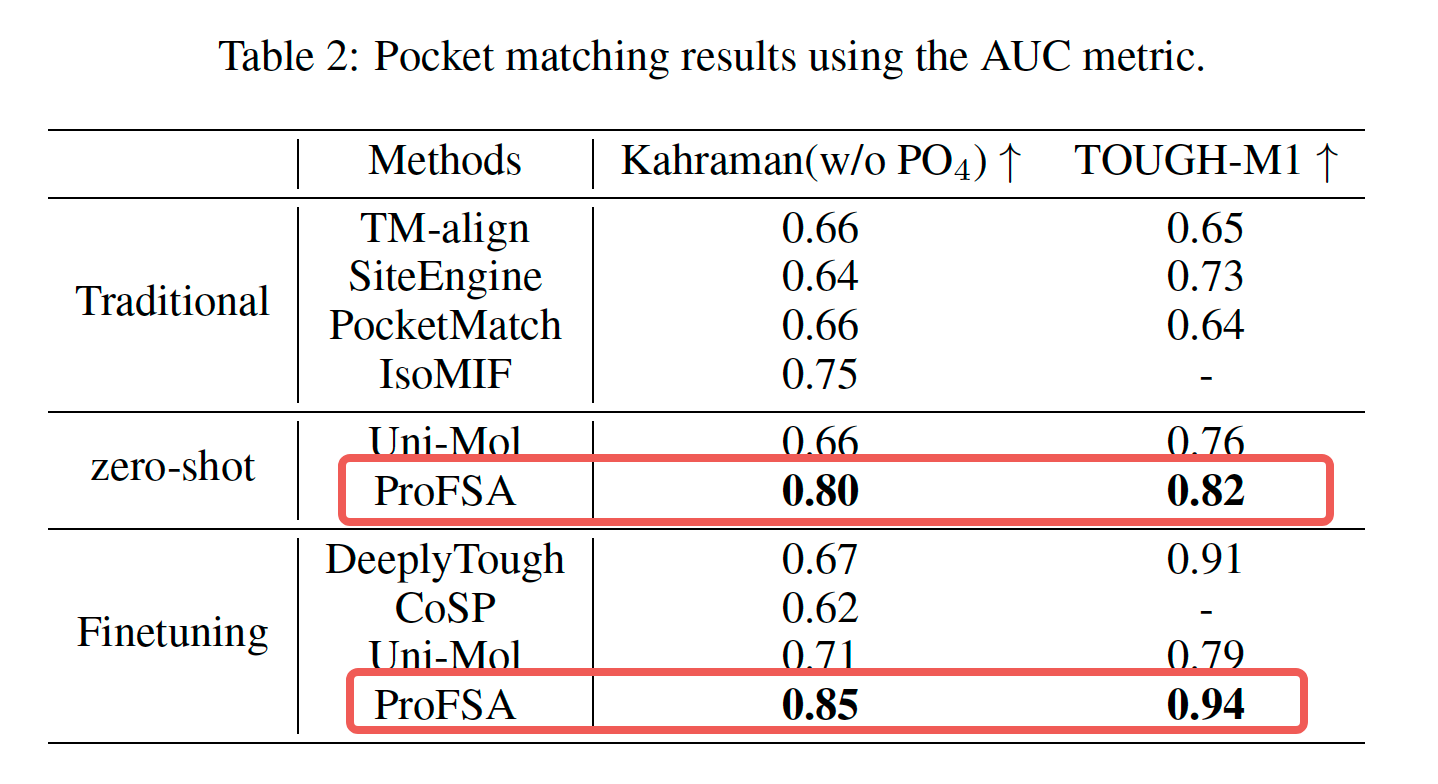
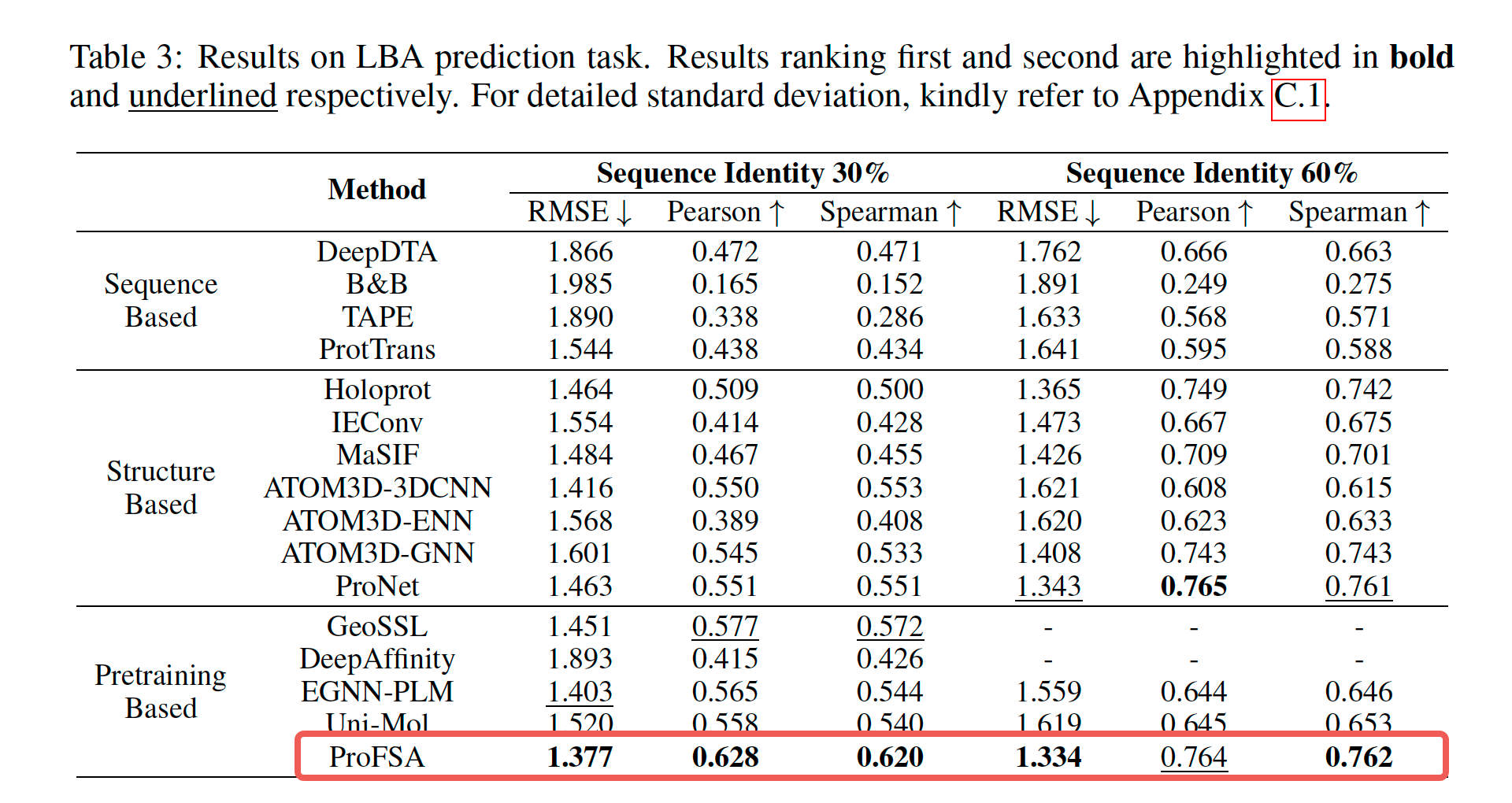
Our method attains state-of-the-art (SOTA) outcomes in experiments involving pocket matching and predicting pocket-ligand binding affinity. Notably, it performs even better when there is a considerable distribution discrepancy between the training and testing datasets.
Data Availability
You can find our processed data at: Google Drive
Each PDB file contain one complex data. R chain is the receptor while the L chain is ligand.