Abstract
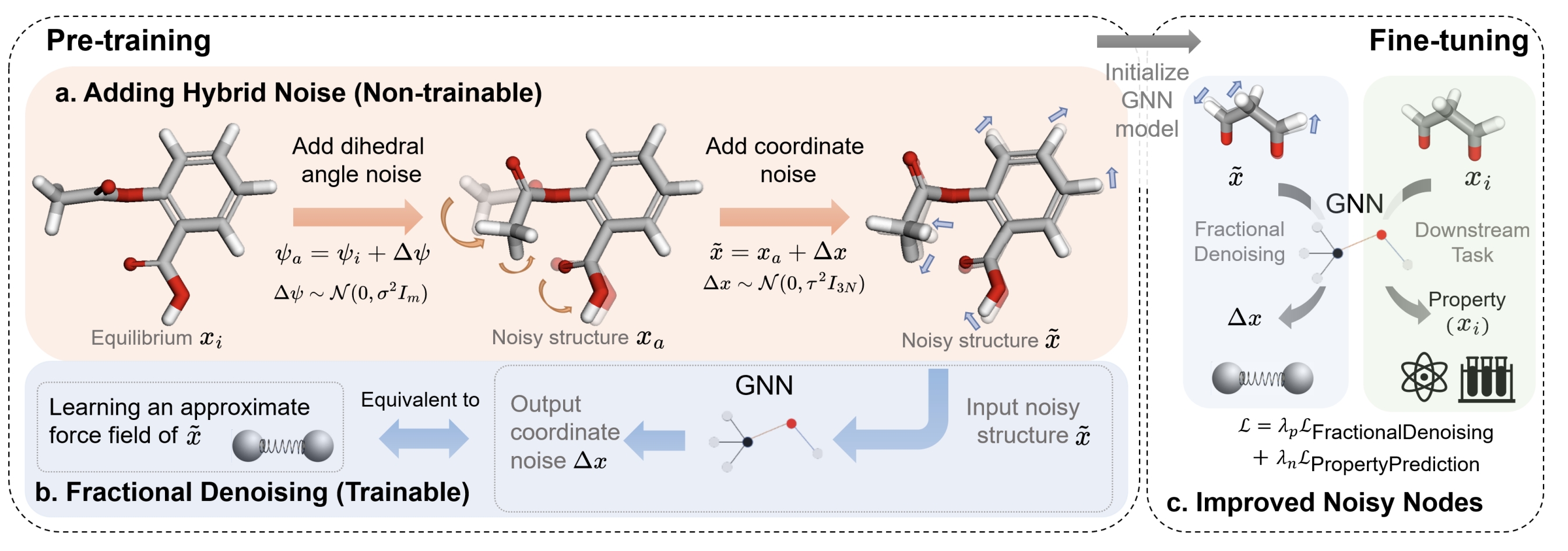
Coordinate denoising is a promising 3D molecular pre-training method, which has achieved remarkable performance in various downstream drug discovery tasks. Theoretically, the objective is equivalent to learning the force field, which is revealed helpful for downstream tasks. Nevertheless, there are two challenges for coordinate denoising to learn an effective force field, i.e. low coverage samples and isotropic force field. The underlying reason is that molecular distributions assumed by existing denoising methods fail to capture the anisotropic characteristic of molecules. To tackle these challenges, we propose a novel hybrid noise strategy, including noises on both dihedral angel and coordinate. However, denoising such hybrid noise in a traditional way is no more equivalent to learning the force field. Through theoretical deductions, we find that the problem is caused by the dependency of the input conformation for covariance. To this end, we propose to decouple the two types of noise and design a novel fractional denoising method (Frad), which only denoises the latter coordinate part. In this way, Frad enjoys both the merits of sampling more low-energy structures and the force field equivalence. Extensive experiments show the effectiveness of Frad in molecular representation, with a new state-of-the-art on 9 out of 12 tasks of QM9 and on 7 out of 8 targets of MD17.
Motivation
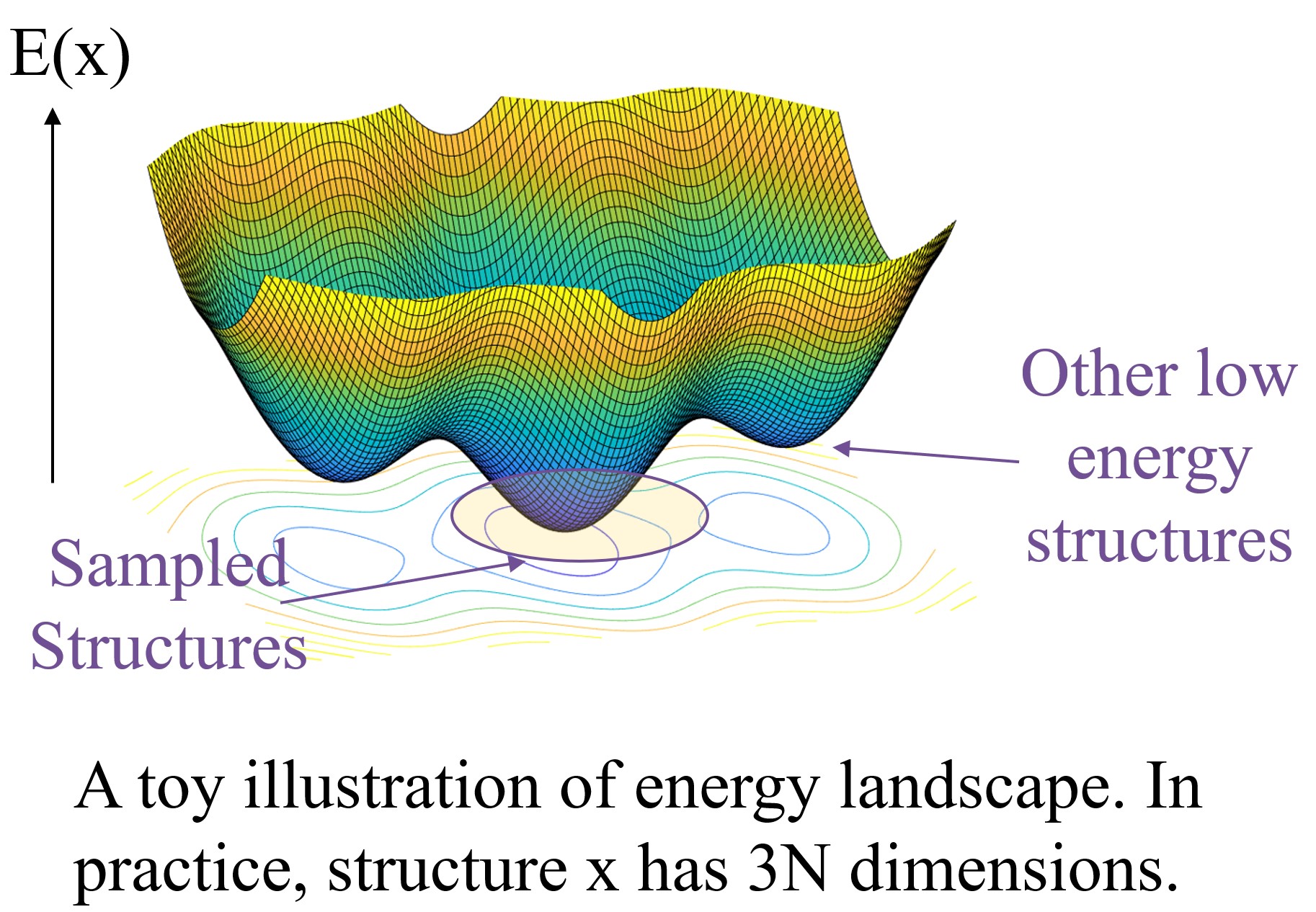
Existing methods suffer from low sampling coverage and inaccurate force field that does not match the anisotropic nature of molecules. Our framework can overcome these problems by refined hybrid noise design to better model the molecular distribution, and maintain the equivalent to force learning by fractional denoising.
Experiments
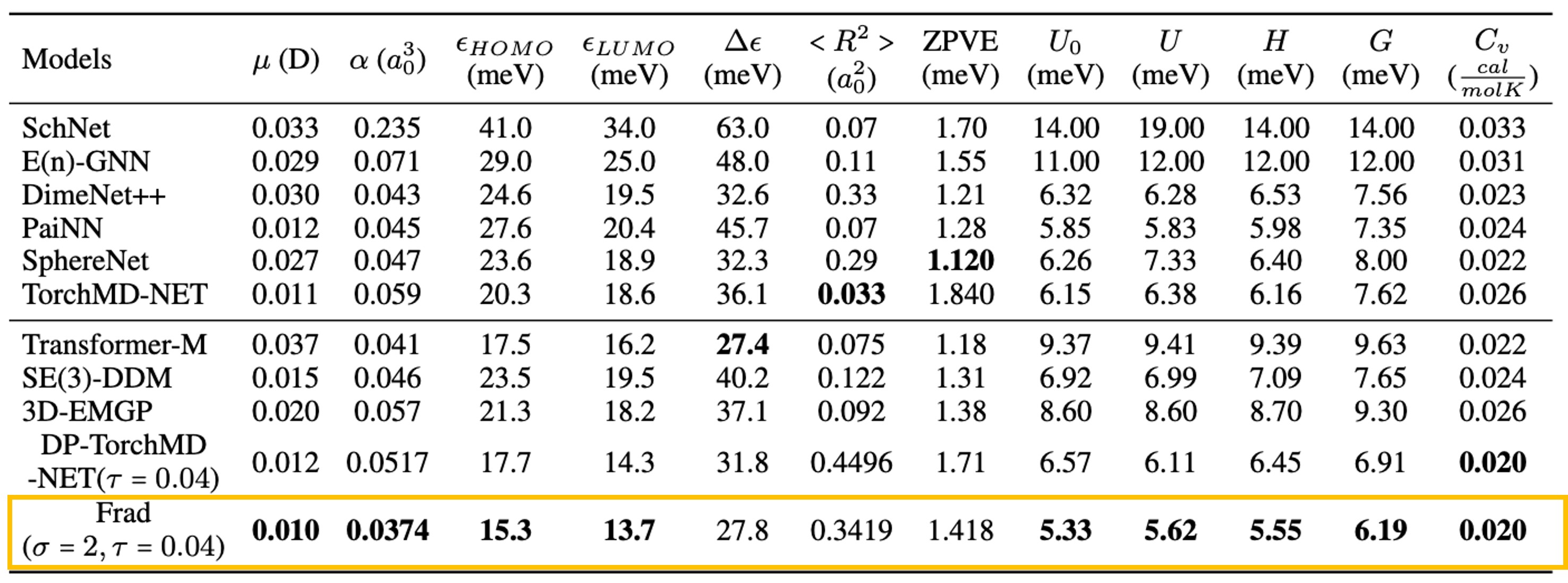
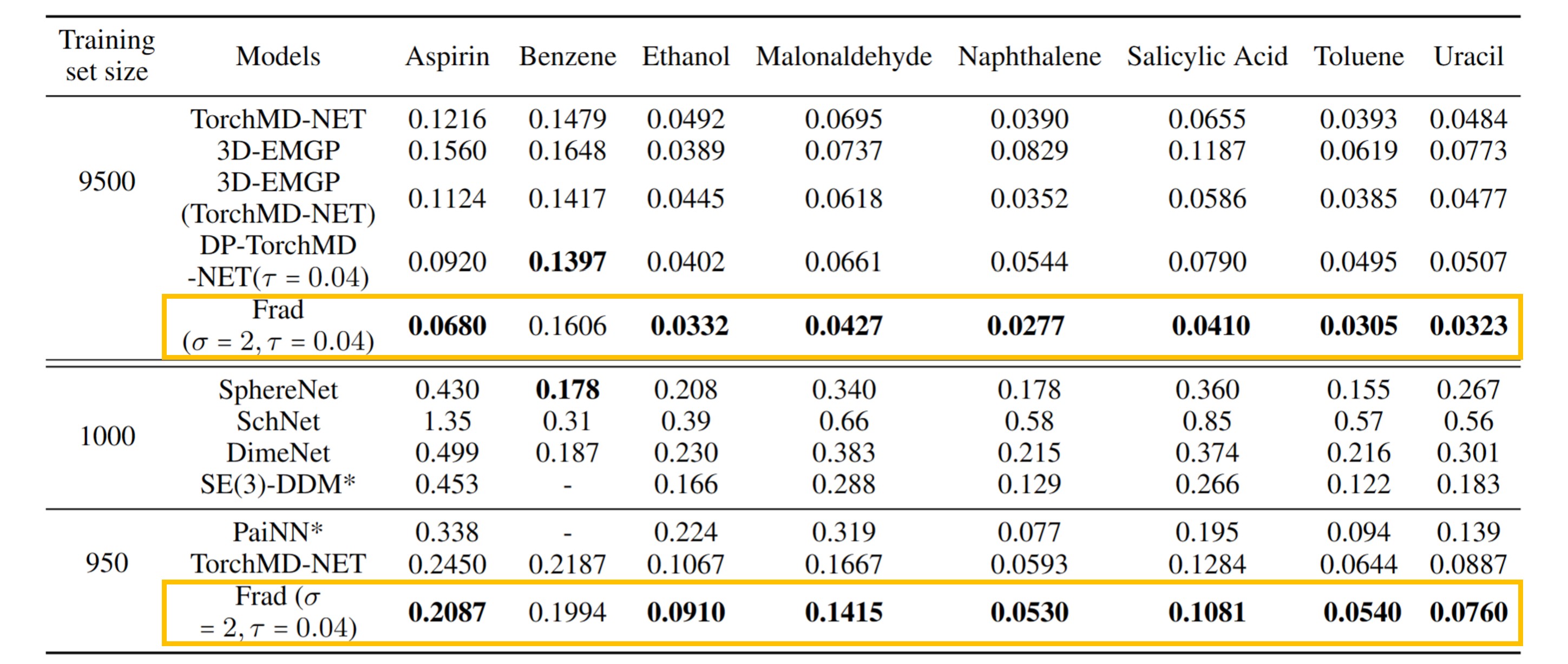
Performances (MAE, lower is better) on QM9 and MD17 force prediction are shown above. The best results are in bold. We new SOTAs on 9/12 tasks of QM9 and 7/8 targets of MD17
BibTex
@InProceedings{pmlr-v202-feng23c,
title = {Fractional Denoising for 3{D} Molecular Pre-training},
author = {Feng, Shikun and Ni, Yuyan and Lan, Yanyan and Ma, Zhi-Ming and Ma, Wei-Ying},
booktitle = {Proceedings of the 40th International Conference on Machine Learning},
pages = {9938--9961},
year = {2023},
volume = {202},
series = {Proceedings of Machine Learning Research},
month = {23--29 Jul},
publisher = {PMLR},
pdf = {https://proceedings.mlr.press/v202/feng23c/feng23c.pdf},
url = {https://proceedings.mlr.press/v202/feng23c.html},
}