Abstract
Virtual screening, which identifies potential drugs from vast compound databases to bind with a particular protein pocket, is a critical step in AI-assisted drug discovery. Traditional docking methods are highly time-consuming, and can only work with a restricted search library in real-life applications. Recent supervised learning approaches using scoring functions for binding-affinity prediction, although promising, have not yet surpassed docking methods due to their strong dependency on limited data with reliable binding-affinity labels. In this paper, we propose a novel contrastive learning framework, DrugCLIP, by reformulating virtual screening as a dense retrieval task and employing contrastive learning to align representations of binding protein pockets and molecules from a large quantity of pairwise data without explicit binding-affinity scores. We also introduce a biological-knowledge inspired data augmentation strategy to learn better protein-molecule representations. Extensive experiments show that DrugCLIP significantly outperforms traditional docking and supervised learning methods on diverse virtual screening benchmarks with highly reduced computation time, especially in zero-shot setting.
Method
Framework
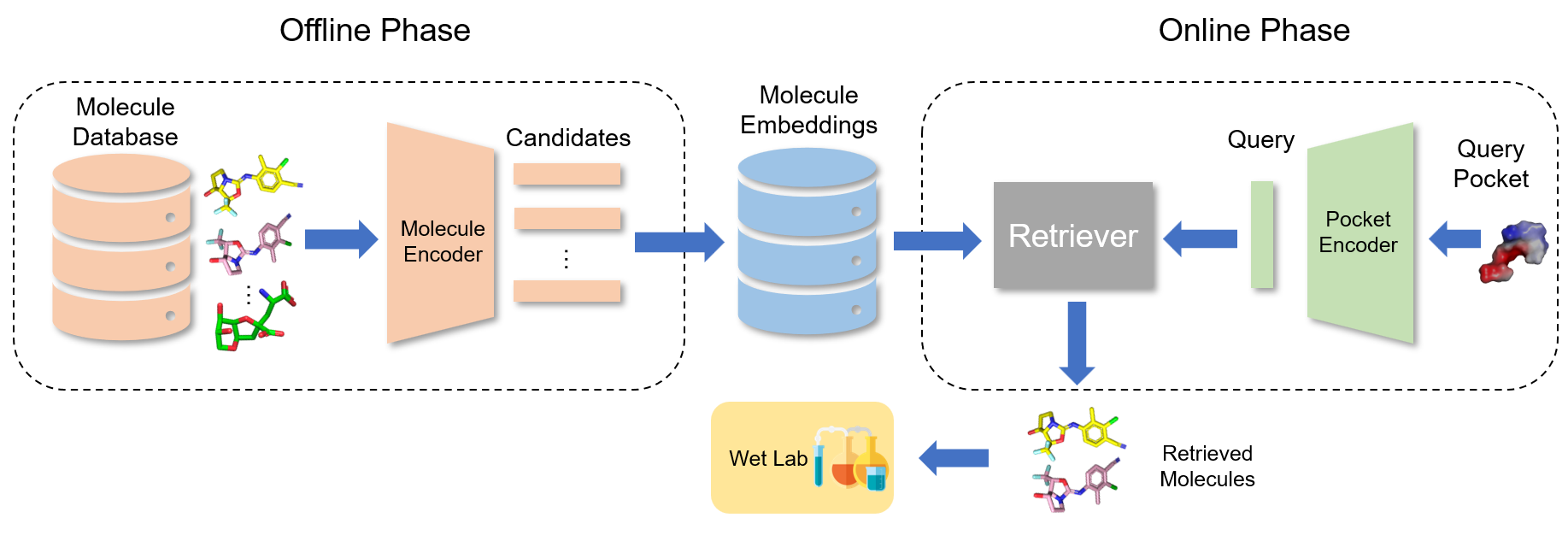
Training
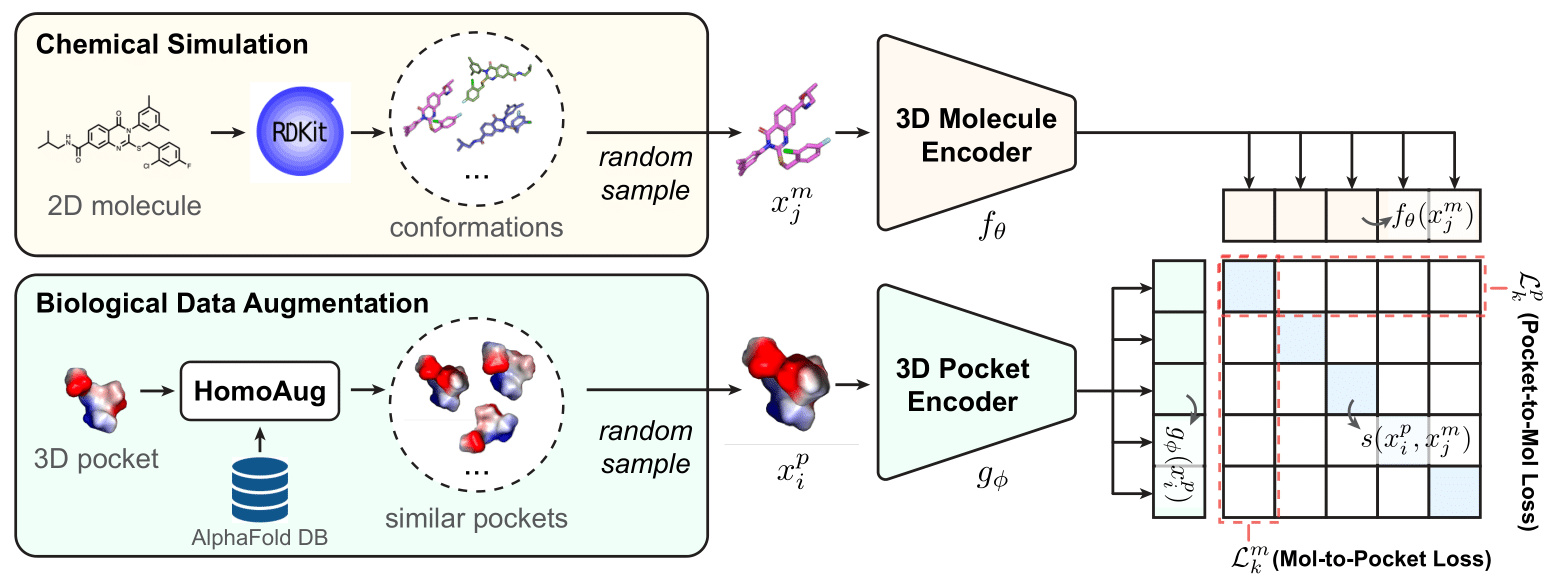
Experiment Results
We assess our methods using two benchmark datasets: DUD-E, which contains topologically diverse decoys with matched physical properties, and LIT-PCBA, a more challenging set that tackles the bias issue prevalent in similar benchmarks. DrugCLIP achieves SOTA results in both cases.
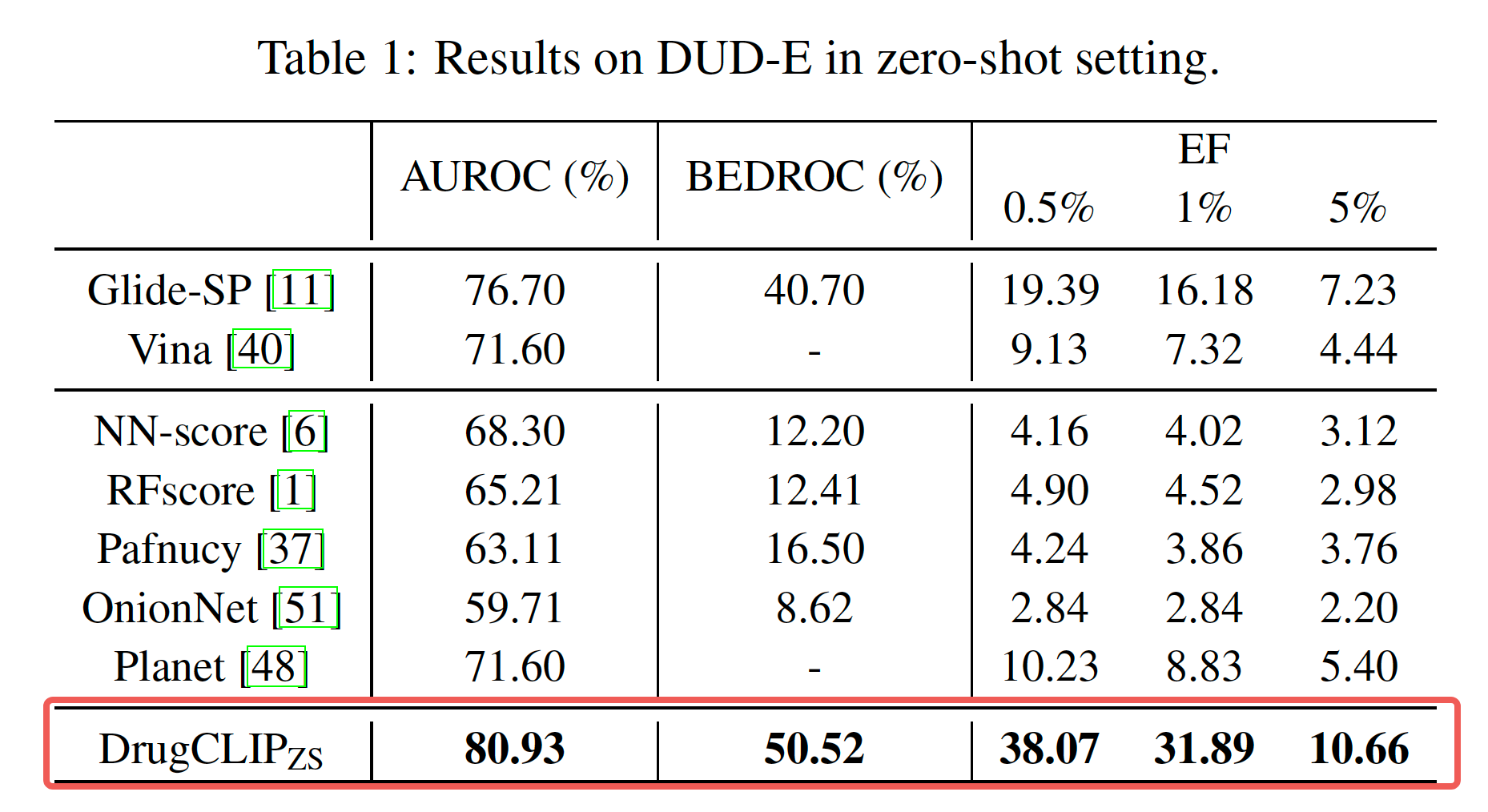
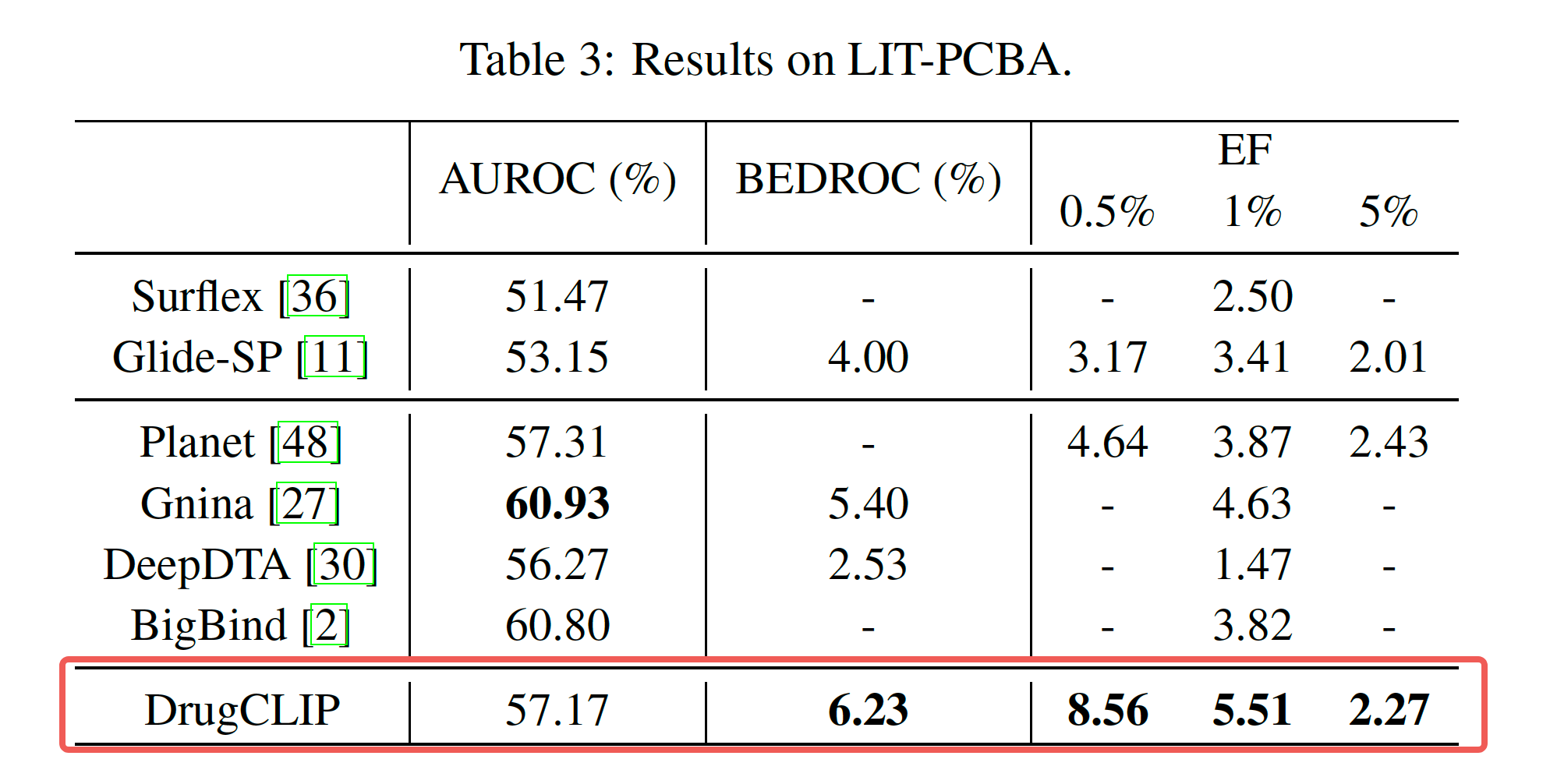
Demo Video
We are in the process of creating a platform that offers pharmacy researchers an intuitive, effective, and efficient method for virtual screening.
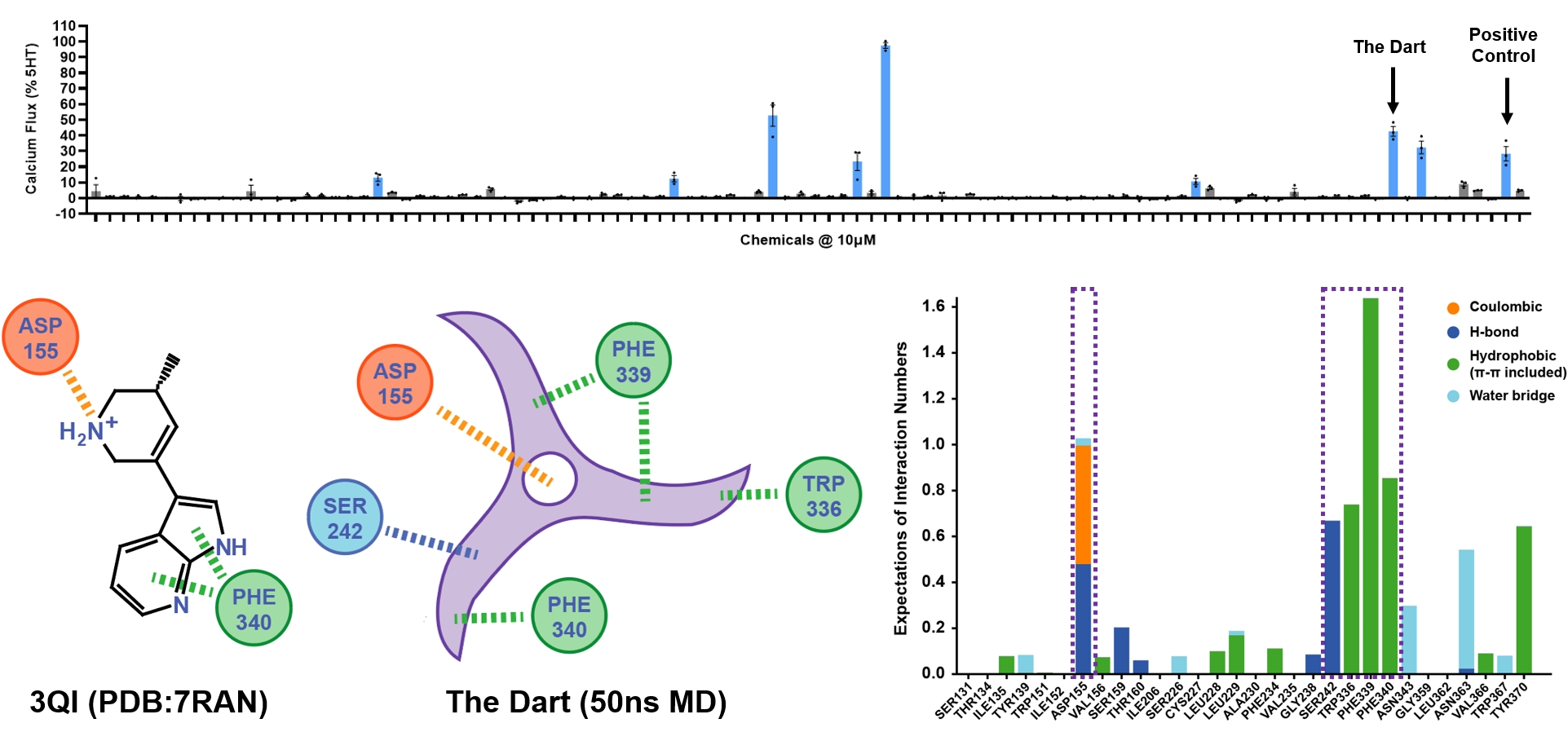
Wet-Lab Results
The 5HT2A receptor plays a vital role in the brain, involved in mood and cognitive regulation, and is thus a significant target for drugs addressing psychiatric conditions like depression, schizophrenia, and anxiety. Our team carried out virtual screening within the ChemDiv molecular library and selected candidates for in vitro testing. The chosen molecules demonstrated over 10% activity at a concentration of 10 micromolar in all three biological replicate experiments.
BibTeX
@inproceedings{gao2023drugclip,
author = {Gao, Bowen and Qiang, Bo and Tan, Haichuan and Jia, Yinjun and Ren, Minsi and Lu, Minsi and Liu, Jingjing and Ma, Wei-Ying and Lan, Yanyan},
title = {DrugCLIP: Contrasive Protein-Molecule Representation Learning for Virtual Screening},
booktitle = {NeurIPS 2023},
year = {2023},
month = {October},
url = {https://www.microsoft.com/en-us/research/publication/drugclip-contrasive-protein-molecule-representation-learning-for-virtual-screening/},
}